Expression Datasets
01 Annona squamosa Pulp Ripening
RNA-Seq of Annona squamosa (cv. Gefner and African Pride) pulp at different stages of growth and ripening.
Experimental Conditions (3 biological replicates):
- African Pride pulp 30 DAP.
- African Pride pulp 100 DAP.
- African Pride pulp 130 DAP.
- African Pride pulp 6 DAH.
- African Pride pulp 8 DAH.
- Gefner pulp 30 DAP.
- Gefner pulp 100 DAP.
- Gefner pulp 130 DAP.
- Gefner pulp 6 DAH.
- Gefner pulp 8 DAH.
The samples were obtained from the "Guangxi Academy of Agricultural Sciences" in Guangxi, China. Thirty fruit samples were collected for each cultivar at five stages of growth and ripening between April and July 2019. Fruits from each cultivar and stage were separated into three biological replicates, and pulp samples were collected, frozen in liquid nitrogen, and used for RNA-seq. The sampled time points correspond to 30, 100, and 130 DAP, as well as 6 and 8 DAH.
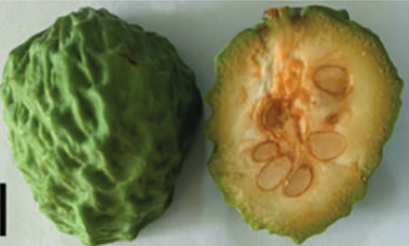







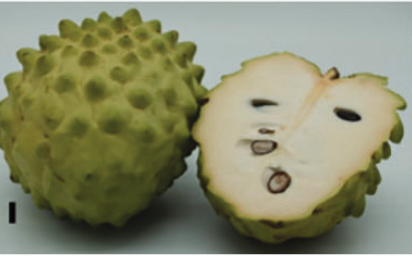

Images of fruits from A. squamosa cv. African Pride (top) and cv. Gefner (bottom) at 30, 100 and 130 DAP; and 6 and 8 DAH.
DAP = Days After Pollination
DAH = Days After Harvest
This dataset was published by Fang et al. 2020, and raw data can be found in the BioProject PRJNA639613.
02 Annona squamosa Flower Development
RNA-Seq of Annona squamosa (cv. Bendi) flowers at different stages of development.
Experimental Conditions:
- Inflorescence meristeme.
- Flower bud.
- Mature flower with partially opened petals.
- Mature flower with opened and faded petals.
The samples were obtained from the "Lingnan Normal University" in Guangdong, China. The inflorescence meristem (IM), the flower buds (FB), and two stages of flowers (FL1 and FL2) were collected from A. squamosa trees. The flower buds were collected based on their size (3–6 mm). The two flower stages were the mature flowers with partially opened petals (FL1) and mature flowers with opened and faded petals (FL2). These tissue samples were frozen immediately in liquid nitrogen, and stored at −80 ºC until use. Equal volumes of RNA from each of the four stages of flower development were pooled, and each sample was prepared by mixing three replicate samples.

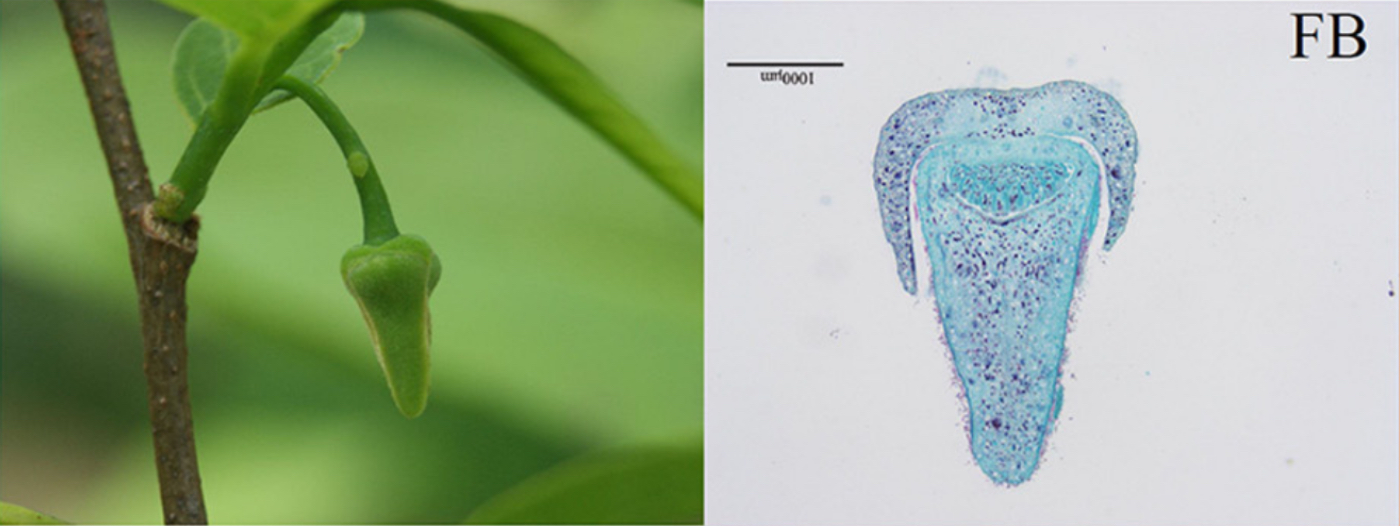
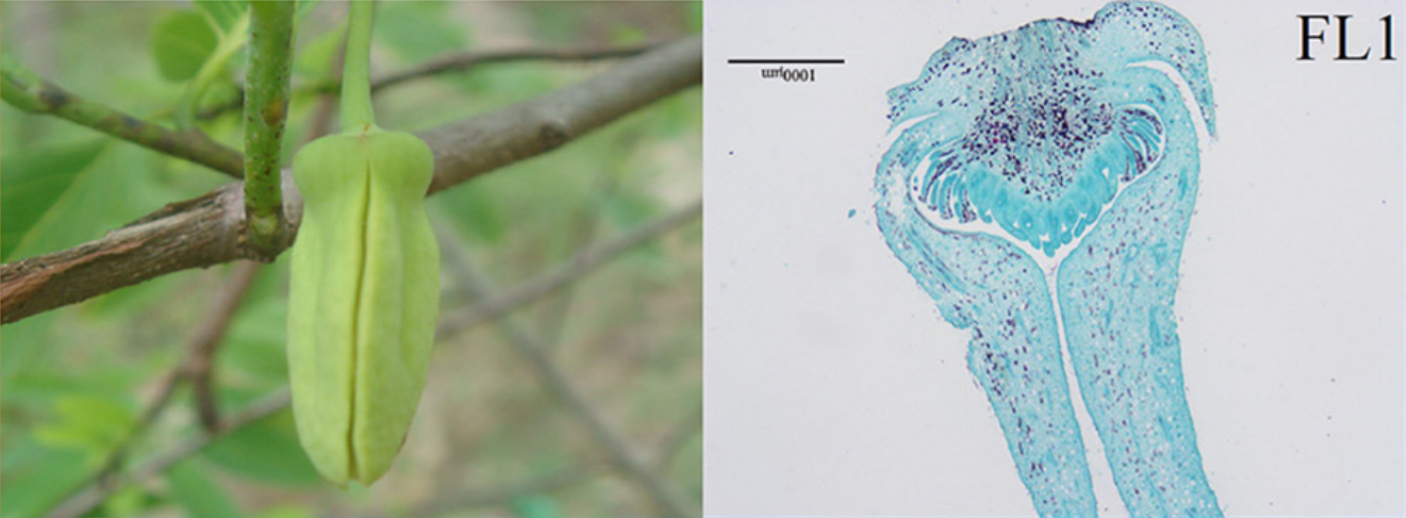
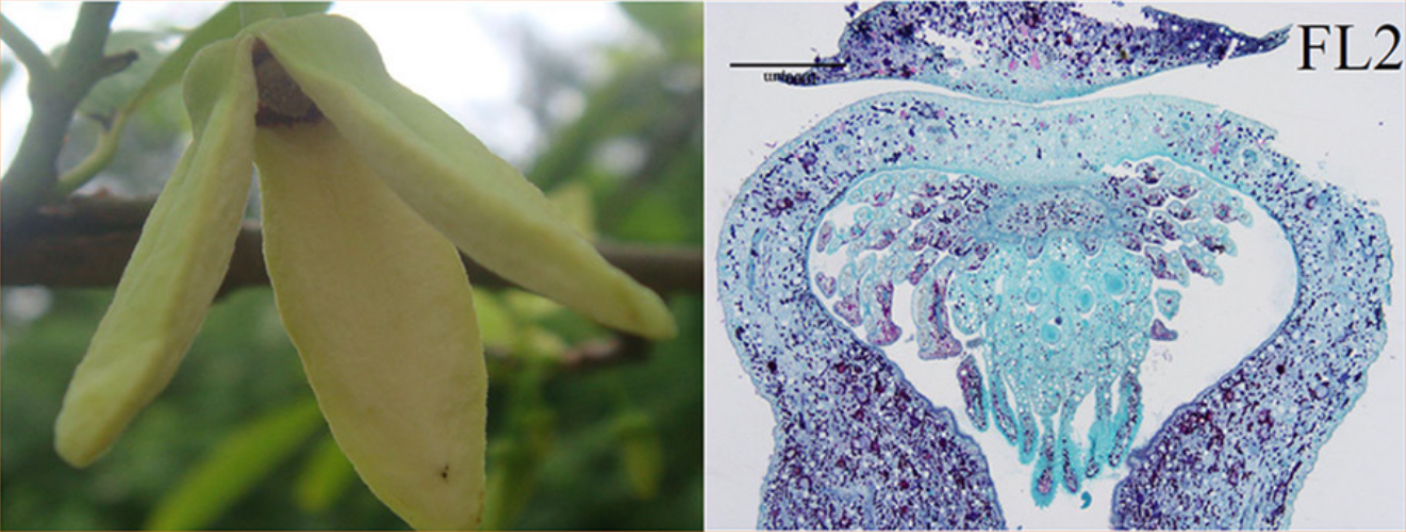
Images of flowers from A. squamosa (cv. Bendi).
IM = Inflorescence Meristeme
FB = Flower Bud
FL1 = Mature flower with partially opened petals
FL2 = Mature flower with opened and faded petals
This dataset was published by Liu et al. 2016, and raw data can be found in the BioProject PRJNA320668.
03 Annona squamosa Normal and Malformed Flowers
RNA-Seq of Annona squamosa (cv. Bendi) normal flowers and malformed flowers.
Experimental Conditions:
- Normal flowers.
- Malformed flowers.
The samples were obtained from the "Lingnan Normal University" in Guangdong, China. The flowers before open were collected from a normal flower and a malformed flower from A. squamosa trees. All of the flower samples were frozen immediately in liquid nitrogen, and stored at −80 ºC until use. Each sample of RNA was prepared by mixing three replicate samples.
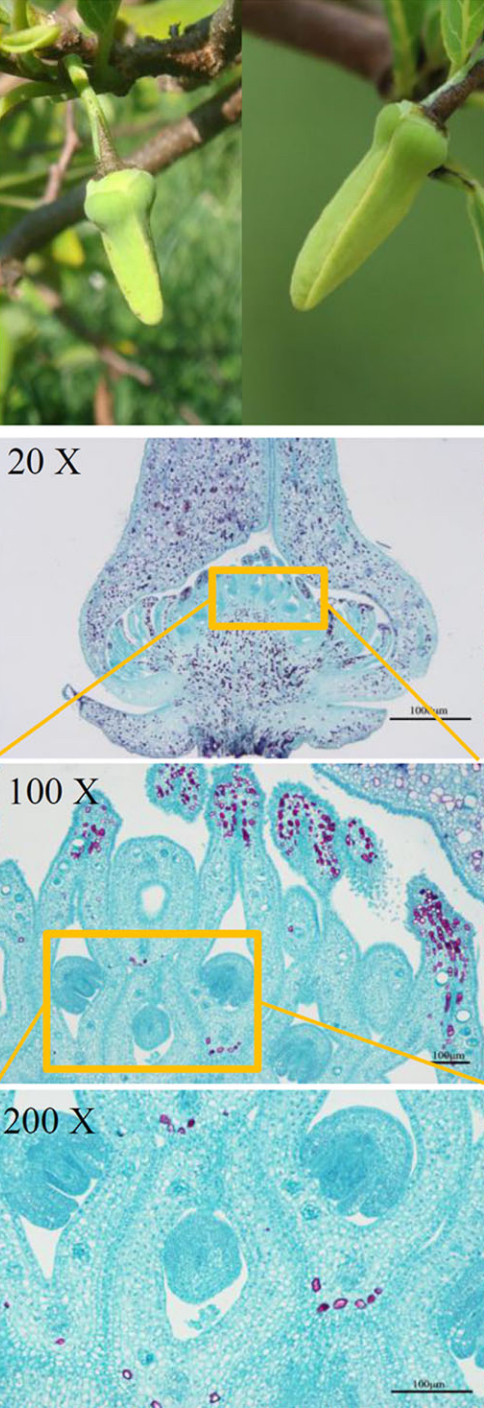
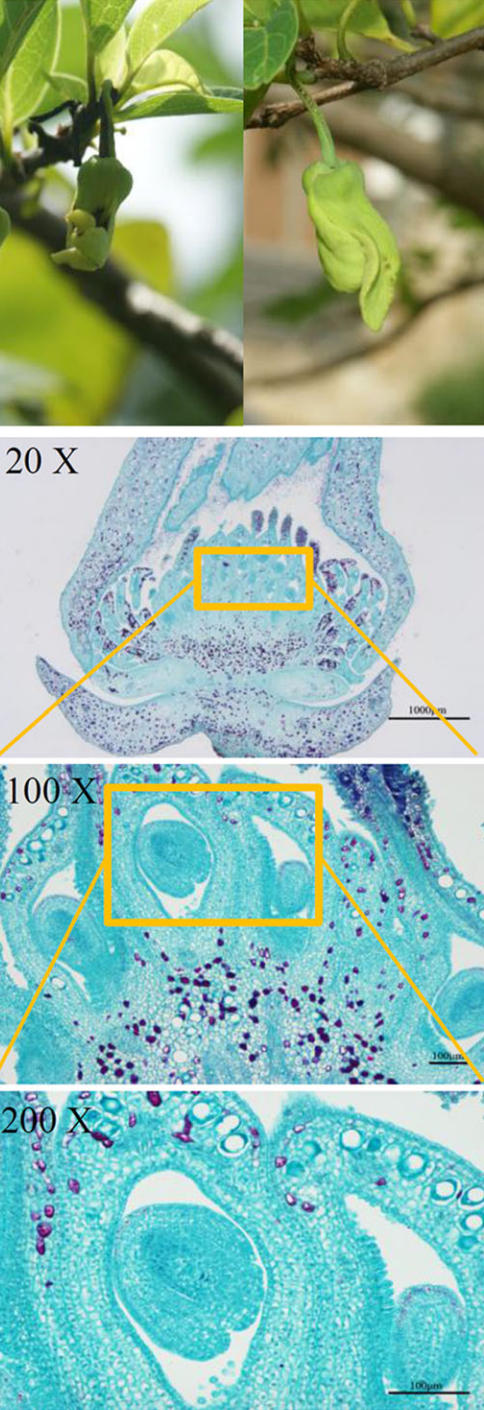
Images of normal flowers (left) and malformed flowers (right) from A. squamosa (cv. Bendi).
This dataset was published by Liu et al. 2017, and raw data can be found in the BioProject PRJNA358456.
04 Atemoya Pericarp Fruits with Ethylene Treatment
RNA-Seq of atemoya (Annona squamosa × A. cherimola) (cv. African Pride) pericarp fruits during post-harvest storage with and without ethylene treatment.
Experimental Conditions:
- Normal treated fruits.
- Ethylene treated fruits.
The samples were obtained from the "Chinese Academy of Tropical Agricultural Sciences" in Guangdong, China. Twenty fruits were kept at room temperature with no treatment, and the following four stages were selected for transcriptome sequencing: recently picked fruits (NT-0), cracking at the pedicel (NT-PC-1), moderate cracking at the pedicel (NT-PC-2), and severe cracking at the pedicel (NT-PC-3). Another twenty fruits were immersed in 2 g/kg ethylene for 2 min, transferred into a closed plastic box, and the lid was left open after 24 hours. The following three stages were selected for transcriptome sequencing: fruits 24 hours after ethylene treatment (Eth-24), fruits with pericarp cracking after ethylene treatment (Eth-PC), and no fruit cracking after ethylene treatment (Eth-NC).
Three similar pericarp samples under the same treatment at each stage were collected, immediately frozen in liquid nitrogen, and then stored at −80 °C until use. Each sample of RNA was prepared by mixing three replicate samples.


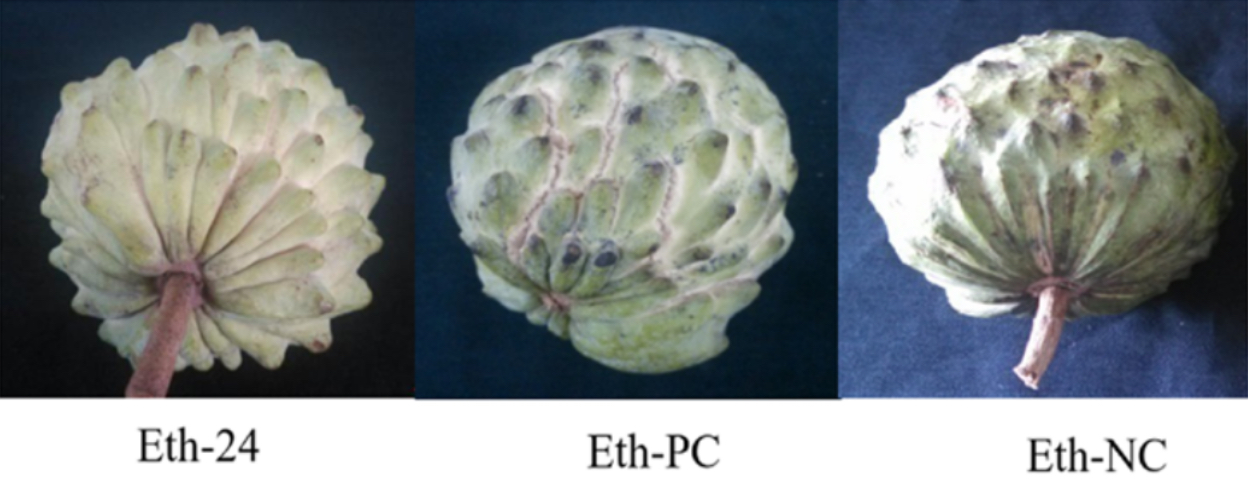
Images of fruits from atemoya (cv. Africa Pride) during ripening and cracking in normal conditions and treated with ethylene.
NT = Normal Treatment
PC = Pericarp Cracking
Eth = Ethylene Treatment
NC = No Cracking
This dataset was published by Chen et al. 2019, and raw data can be found in the BioProject PRJNA396469.
05 Atemoya Split and Non-Split Fruits
RNA-Seq of atemoya (Annona squamosa × A. cherimola) pulp of split and non-split fruits.
Experimental Conditions:
- Split fruits.
- Non-split fruits.
The samples were obtained from the "Lingnan Normal University" in Guangdong, China. All fruits were harvested at 6 months after pollination. Non-split fruits (NSF) and split fruits (SF) were selected at 10 days after harvest. All of the fruit samples were frozen immediately in liquid nitrogen, and stored at −80 ºC until use. Each sample of RNA was prepared by mixing three replicate samples.
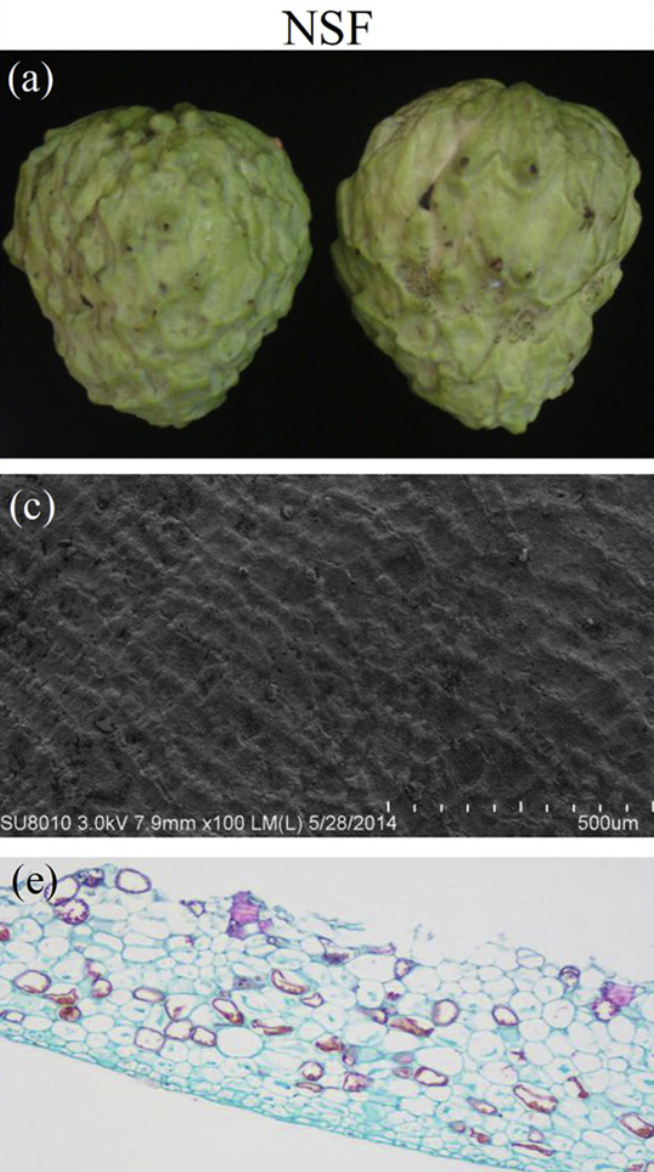
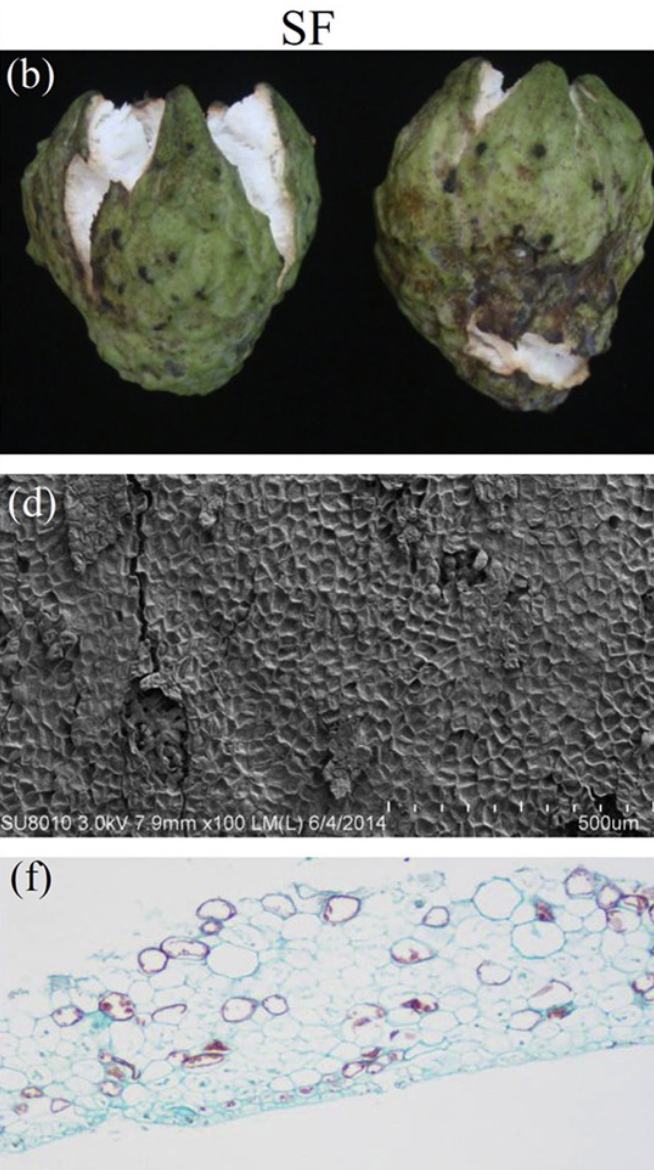
Images of NSF (a) and SF (b) from atemoya. Observation with Scanning Electron Microscopy (SEM) the NSF (c) and SF (d) in atemoya, and observation with Optical Microscope the NSF (e) and SF (f).
NSF = Non-Split Fruits
SF = Split Fruits
This dataset was published by Li et al. 2019, and raw data can be found in the BioProject PRJNA397365.
06 Annona Outer Integument
RNA-Seq of pistils with and without early seed development in Atemoya (Annona squamosa × A. cherimola) cv. Gefner, and the A. squamosa "Thai seedless" mutant (Annona ino).
Experimental Conditions:
- AG = Atemoya cv. Gefner
- T = "Thai seedless" mutant
- PS = Pistils containing early developing seeds (Pollinated pistils with seed).
- P = Pistils without early developing seeds (Pollinated pistils, seed removed).
Total RNA was extracted from pollinated pistils containing early seeds (PS) and pistils without early seeds (P) of Annona cherimola cv. Gefner (AG) and the "Thai seedless" mutant (T), combining the CTAB protocol by Doyle and Doyle, 1987 and the Qiagen RNeasy Plant Mini Kit (Qiagen).
PS and P samples were collected from three independent fruits at 4 and 8 days after pollination (DAP).
Microscopic observations were performed at 8 DAP to illustrate early embryo and nucellus development.
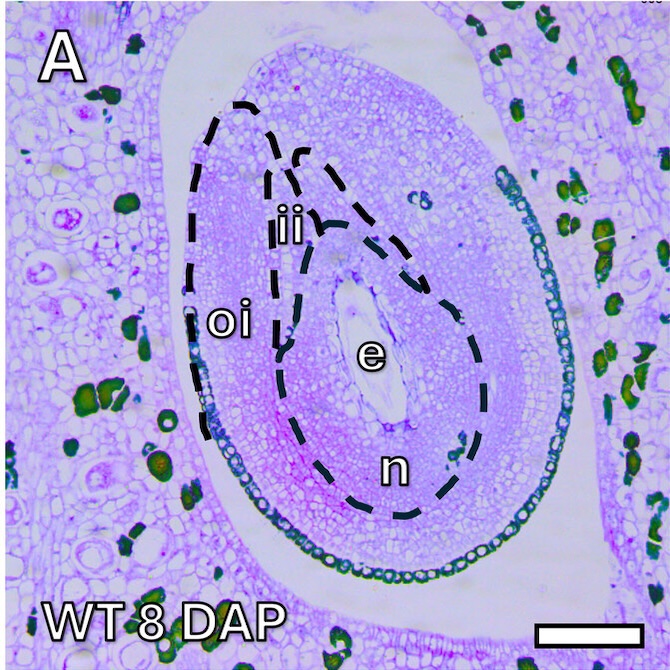
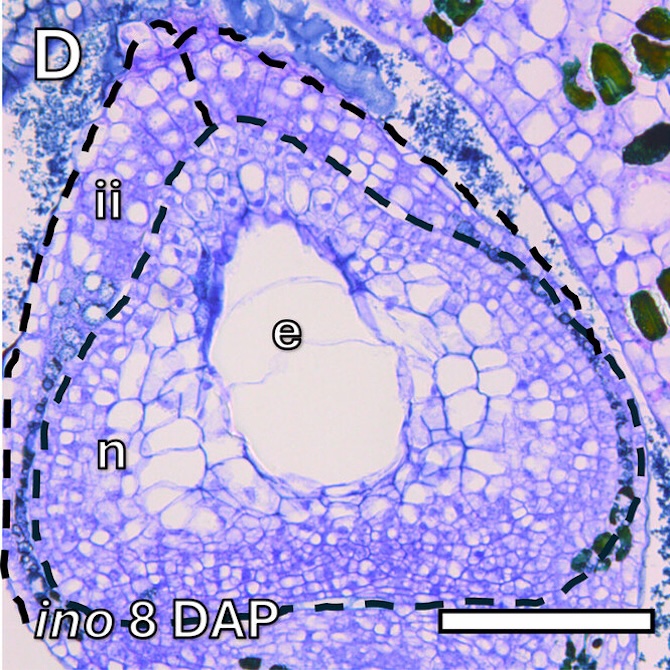
Microscope images of the immature seed sections (8 DAP) for both AG (A) and T (D) genotypes.
e = endosperm
ii = inner integument
n = nucellus
oi = outer integument
Scale bars: 100 μm
This dataset was published by Garcia-Lezama et al., 2025 in Physiologia Plantarum. Raw sequencing data are available in the BioProject PRJEB97455.
Asimina triloba reproductive incompatibility
RNA-Seq of pawpaw (Asimina triloba) early developing seeds collected from three cross-compatible (CC) and three auto-incompatible (AI) pollinated flowers at 4, 8 and 15 days after pollination (DAP), resulting in a total of 18 biological replicates. The pistils of each flower were dissected to obtain the developing seeds, and extract RNA from these fresh early seeds.
Experimental Conditions:
- Developing seeds of cross-compatible pollinated flowers at 4 DAP (CC4)
- Developing seeds of auto-incompatible pollinated flowers at 4 DAP (AI4)
- Developing seeds of cross-compatible pollinated flowers at 8 DAP (CC8)
- Developing seeds of auto-incompatible pollinated flowers at 8 DAP (AI8)
- Developing seeds of cross-compatible pollinated flowers at 15 DAP (CC15)
- Developing seeds of auto-incompatible pollinated flowers at 15 DAP (AI15)
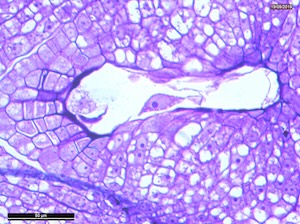
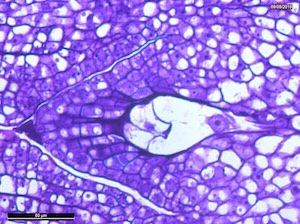 4 DAP
4 DAP
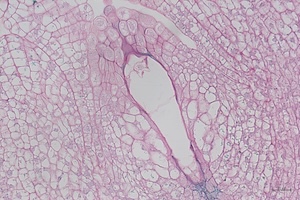
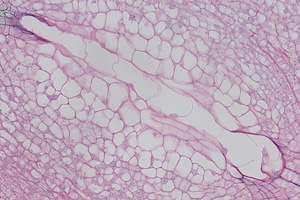 8 DAP
8 DAP
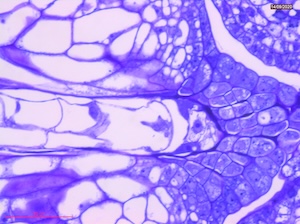
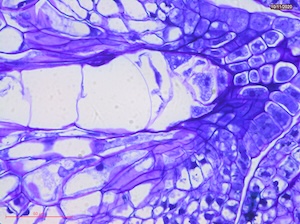 15 DAP
15 DAP
Microscope images of developing seeds of cross-compatible (on the left) and auto-incompatible crosses (on the right).
CC = compatible crosses
AI = Autoincompatible crosses
DAP = Days After Pollination
This dataset was published by Ferrer-Blanco et al. 2025, and raw data can be found in the BioProject PRJEB97068.
T.cacao Phytophthora infection
RNA-Seq of pods of seven cacao genotypes (4 susceptible: ICS1, WFT, Gu133, Spa9, and 3 resistant: CCN51, Sca6 and Pound7) to study their response to the post-penetration stage of Phytophthora palmivora infection.
| Variety | Genetic Background / Characteristics |
|---|---|
| Pound7 | Upper Amazon Forastero lineage; known for specific disease resistance traits. |
| Sca6 | Scavina 6; a foundational clone for Crinkly Leaf and Black Pod resistance. |
| CCN51 | Colección Castro Naranjal; a high-yielding, widely cultivated commercial hybrid. |
| Spa9 | Selection from the 'Servicio de Plantas Agrícolas' (Ecuador). |
| Gu133 | Guiana selection; representing wild genetic diversity from French Guiana. |
| WFT | Specific selection used for pathology and transcriptomic modeling. |
| ICS1 | Imperial College Selection 1; a high-quality Trinitario clone. |
Experimental Conditions (3 replicates):
- Control Group: Healthy pod tissues, non-infected.
- Infection Group: Pod tissues infected with Phytophthora palmivora, the primary causal agent of Black Pod disease.
Methodology & Bioinformatics Pipeline:
- Library Generation: RNA-Seq libraries were generated specifically from cacao pod RNA.
- Platform: Sequencing was conducted on an Illumina platform.
- Mapping: Reads were aligned to the reference Matina v2 genome.
- Quantification: Gene expression levels were normalized using TPM (Transcripts Per Million) values to allow for accurate comparative analysis across genotypes and treatments.
This dataset was published by Baruah et al. 2024 and raw data can be found in the BioProject PRJNA487154.


